
MedSAM2: Segment Anything in 3D Medical Images and Videos
1AI Collaborative Centre, University Health Network, Toronto, Canada
2Vector Institute for Artificial Intelligence, Toronto, Canada
3Department of Biomedical Informatics, Harvard Medical School, Harvard University, Boston, USA
4Peter Munk Cardiac Centre, University Health Network, Toronto,Canada
5Department of Computer Science, University of Toronto, Toronto, Canada
6Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, Canada
7Roche Canada and Genentech
Highlights
- A promptable foundation model for 3D medical image and video segmentation
- Trained on 455,000+ 3D image-mask pairs and 76,000+ annotated video frames
- Versatile segmentation capability across diverse organs and pathologies.
- Extensive user studies in large-scale lesion and video datasets demonstrate that MedSAM2 substantially facilitates annotation workflows
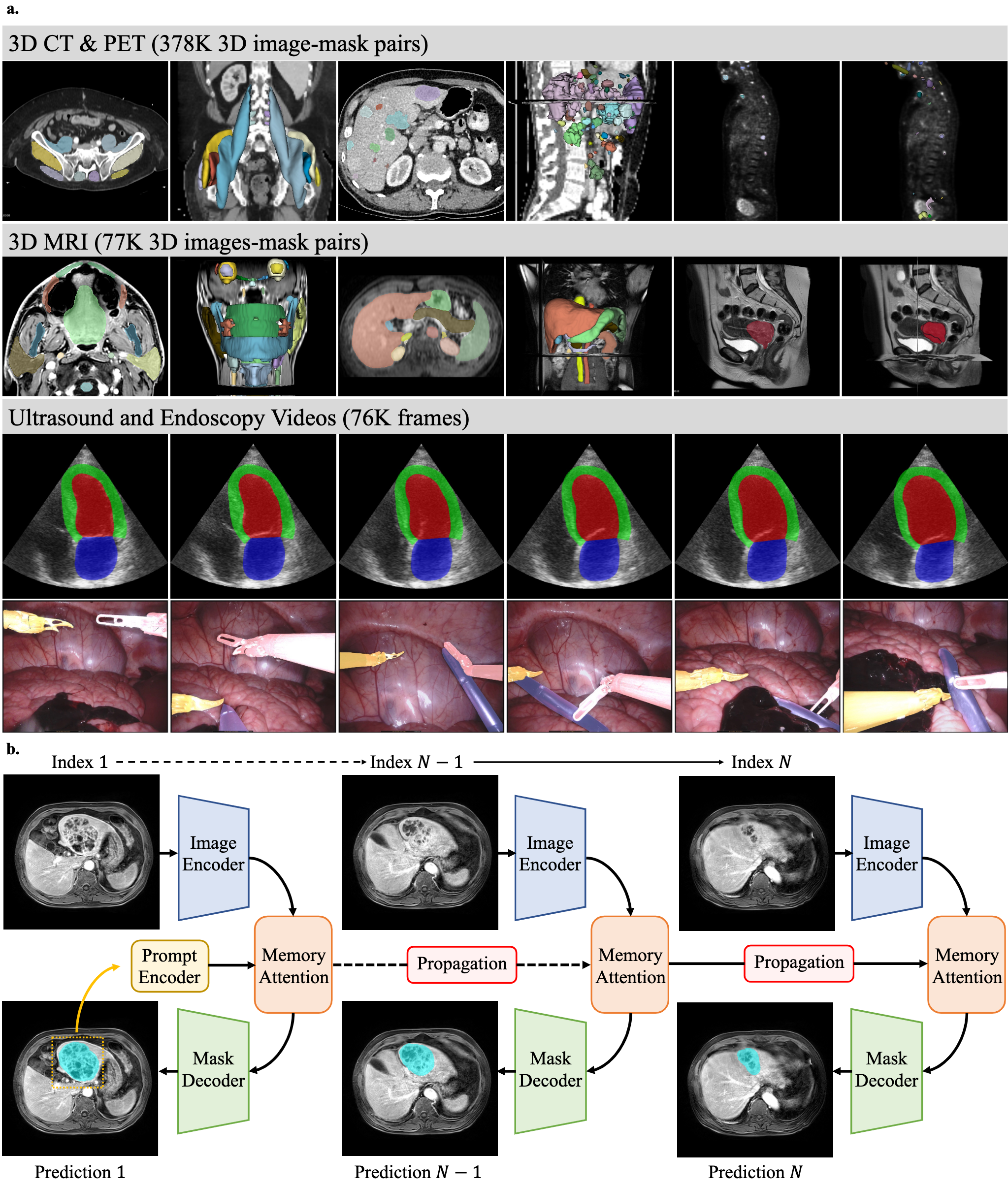
The dataset includes diverse 3D CT, PET, MRI images, ultrasound, and endoscopy videos. For each 3D image example, we visualize both 2D slices and 3D structures. For each video example, we visualize frames at different time points. MedSAM2 is a promptable segmentation network with an image encoder, a prompt encoder, a memory attention module, and a mask decoder. The image encoder extracts multiscale features from each frame or 2D slice. The memory attention module conditions the current frame features on past frames' features and predictions using streaming memory. The mask decoder generates accurate segmentation masks based on bounding box prompts and memory-conditioned features. This architecture enables MedSAM2 to effectively segment both 3D medical images and videos by exploiting spatial continuity across slices and frames.
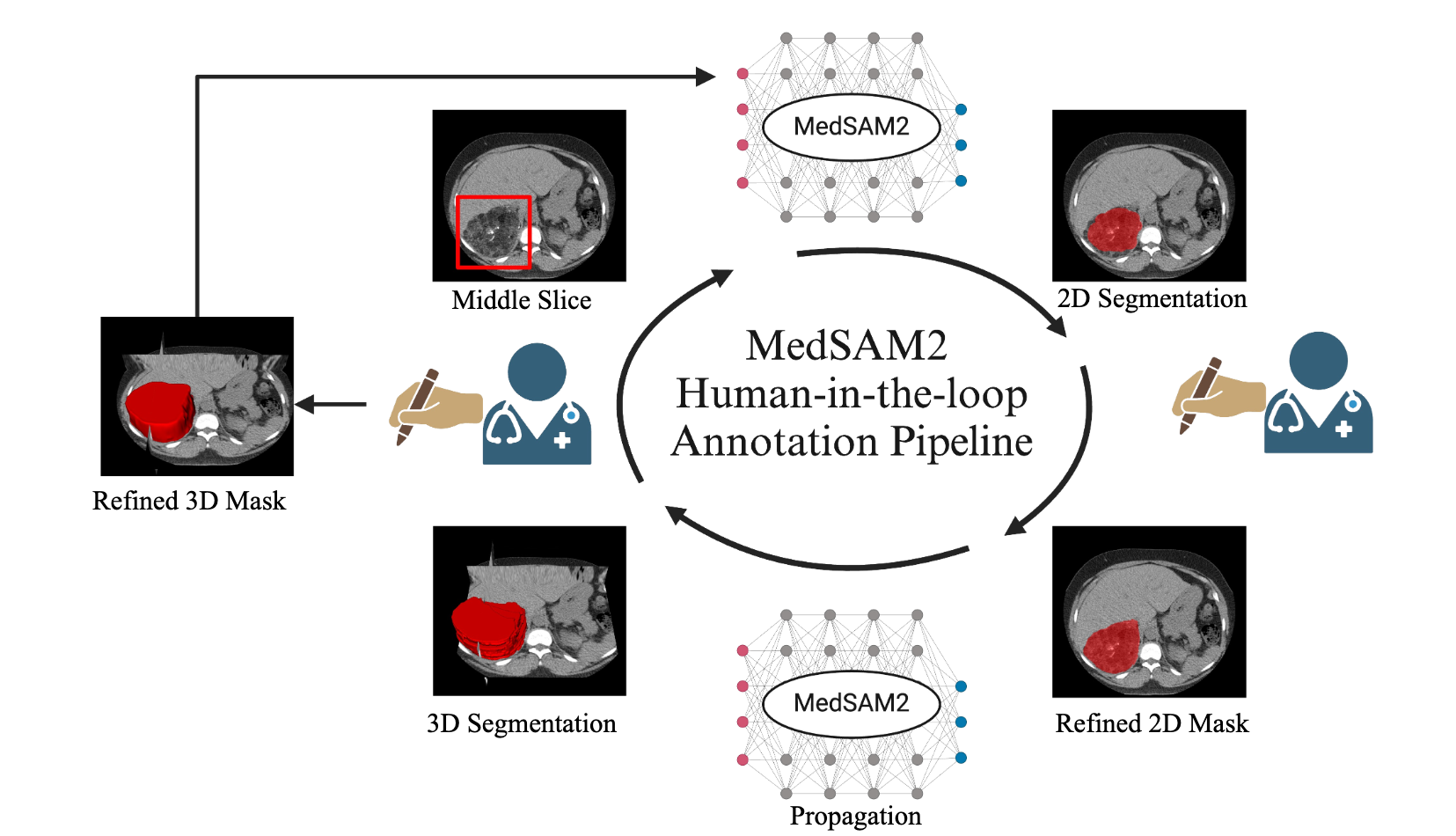
Human-in-the-loop Annotation Pipeline with MedSAM2
MedSAM2 Deployment on Common Computing Platforms
BibTeX
@article{MedSAM2,
title={MedSAM2: Segment Anything in 3D Medical Images and Videos},
author={Ma, Jun and Yang, Zongxin and Kim, Sumin and Chen, Bihui and Baharoon, Mohammed and Fallahpour, Adibvafa and Asakereh, Reza and Lyu, Hongwei and Wang, Bo},
journal={arXiv preprint arXiv:2504.03600},
year={2025}
}
Credit: This website template is adapted from here.









